How bacteria grow: evolutionary differences point to new ways to combat infection

Closely related bacterial species use different strategies to build their cell walls, an international team of scientists led by Université de Montréal microbiology professor Yves Brun has found.
Their discovery promises to help guide new approaches in the fight against antibiotic resistance, they say in their research, published in June in Nature Communications.
With colleagues from his department as well as the Institut national de la recherche scientifique and Indiana University, Brun's research team used advanced microscopy techniques and fluorescent probes to track bacterial growth.
The results reveal an unexpected diversity in wall elongation patterns, centered on peptidoglycan synthesis – an important target of several classes of antibiotics, including penicillin.
“What was thought to be a relatively uniform process actually appears to be much more variable, even between very closely related species,” said Brun, the study's lead author. “This variability may represent new points of fragility that could be exploited to develop new antibiotics.”
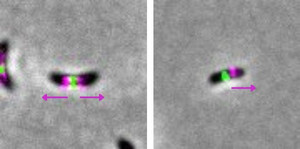
The study focused on the Caulobacteraceae family. The researchers compared Caulobacter crescentus, a model organism, with Asticcacaulis excentricus, a related species. While the former elongates its wall bidirectionally from the center, the latter does so only towards one cell pole. This change is associated with a relocation of the PBP2 enzyme, a classic target of antibiotics.
But that's not all. “It was while observing this unexpected evolutionary diversity between closely related species that we had the idea of testing whether elongation modes could also vary according to environment,” explained co-first author Marie Delaby, a postdoc in Brun's lab.
"And indeed, new data we've obtained since the submission of the article (to Nature Communications last November) show that these mechanisms are not only determined by genetics, they can also adjust to environmental conditions," she said.
"This shows that the study of evolution doesn't just serve to reconstruct the history of bacteria. It can also reveal little-known forms of variability, which is useful for better understanding and potentially countering antibiotic resistance."
The observations by Burn's team suggest that some bacteria may actively modulate their growth according to context, complicating the action of current treatments. By taking this plasticity into account, the researchers hope to contribute to the development of better adapted strategies for targeting bacteria in different environments.
“We need to stop thinking of bacterial wall synthesis as a fixed, universal target,” said Brun. "It's a more dynamic process than we thought, influenced by evolution and the environment. This realization can enrich our approaches to the development of new treatments."



